Introduction
A best practices approach to single-cell analysis requires combining methods from the three major ecosystems: scverse for Python and Bioconductor or Seurat for R. Each of these ecosystems has its own core object for storing single-cell data with its own structure and interface. To take advantage of the strengths of each ecosystem, analysts need to be able to efficiently move data between them which can require detailed knowledge of the different data structures. Moving data between Python and R presents an additional challenge which can often limit analysts to using one language.
Started at the scverse hackathon in 2023, anndataR solves this problem by providing an R-native interface for H5AD files and unlocking the defacto single-cell file format for R users. This post will introduce the key features of anndataR but more detail is available in the preprint.
anndataR addresses key challenges of interoperability between R and Python
Several approaches for transferring single-cell data already exist, many of which rely on foreign function interfaces (FFIs) such as reticulate or rpy2. Using an FFI has advantages, it allows data to be moved interactively between concurrent sessions of the two languages and avoids reimplementing functionality. However, it also presents difficulties as the user must set up and manage environments for both languages. This can be worthwhile when using a complex method from a foreign language but can be a significant overhead when you simply want to transfer data.
The approach taken by anndataR is to provide R-native reading and writing of H5AD files, conforming to the official specification. The HDF5-based H5AD format developed by the scverse community has become the de facto standard for storing single-cell data on disk and anndataR allows R users to access data in this format without needing to manage a Python installation.
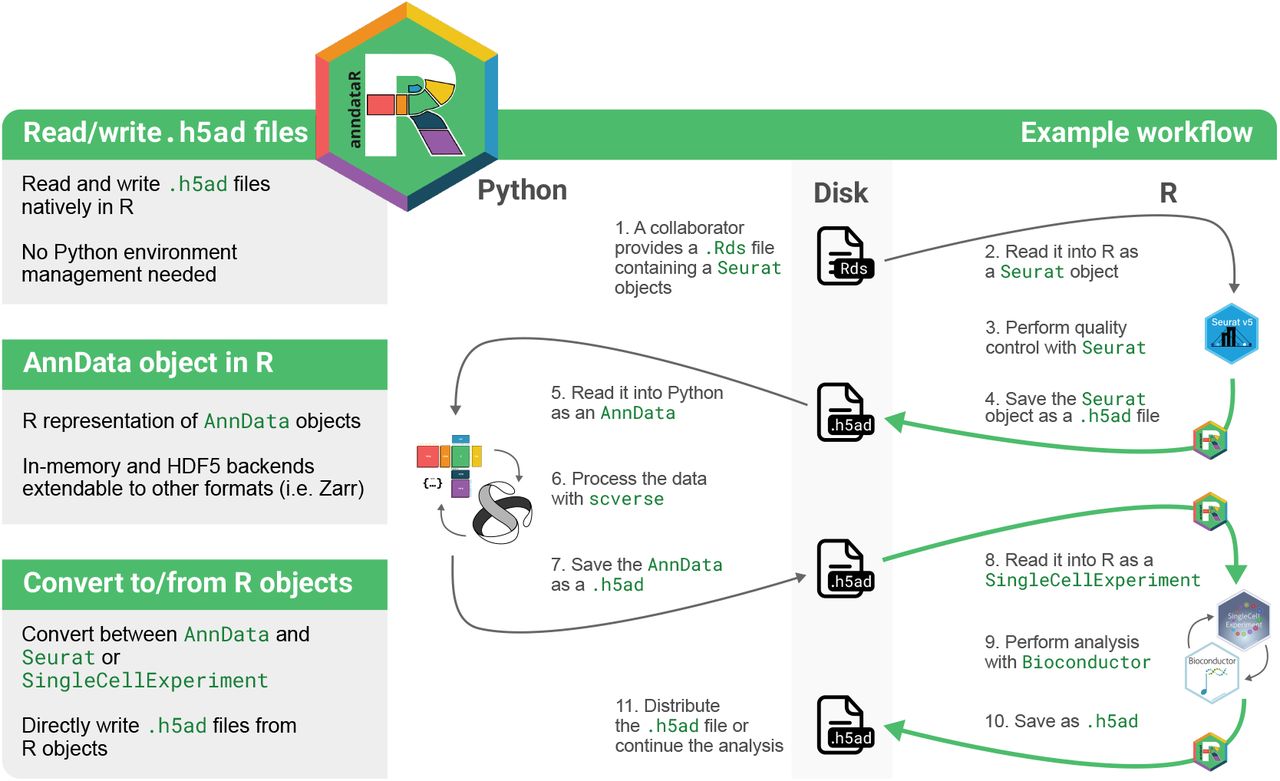
Key features of anndataR
- Native reading and writing of H5AD files, conforming to the on-disk AnnData specification
- Direct conversion to and from Seurat or SingleCellExperiment objects
- Flexible control over which items are converted, allowing partial reading of H5AD files
- In-memory and H5AD-backed R implementations of the AnnData object
Future work will include reading and writing of the AnnData Zarr format, support for lazy disk-backed access and additional functionality for the AnnData object.
Ensuring robust compatibility between R and Python
Developing a new implementation of an existing standard opens the door to a range of compatibility issues. Designing anndataR to match the AnnData on-disk specification helps to minimise these but is not sufficient by itself. To fully ensure compatibility with the Python implementation there is a comprehensive suite of over 1800 unit tests for anndataR, including many roundtrip tests. When a known incompatibility is found, these are listed on the anndataR website to notify users and so they can be addressed during further development.
The close collaboration between the anndataR developers and the scverse core team also allows coordination on any future changes to the AnnData format.
Thank you to everyone who contributed to anndataR!
The anndataR package is a community project and wouldn’t be possible without everyone who contributed:
- All the developers who contributed code to the package, particularly co-lead developer Louise Deconinck
- The scverse core team for organising the kick-off hackathon and their support since then
- Funding from the Chan Zuckerberg Initiative (CZI) through the Essential Open Source Software for Science grant EOSS6-000000074
We welcome further contributions from the community through GitHub!
Links
- GitHub https://github.com/scverse/anndataR
- Website https://anndatar.data-intuitive.com/
- Preprint https://doi.org/10.1101/2025.08.18.669052
- Bioconductor submission https://github.com/Bioconductor/Contributions/issues/3907
