Dynverse, utilizing the ‘dyno’ package, is a state-of-the-art framework designed for inferring cellular trajectories from single-cell RNA sequencing data. It offers a comprehensive suite of tools for robust and efficient trajectory analysis, ensuring high reproducibility and comparability across methods. Dynverse facilitates the exploration of cellular dynamics, aiding in the understanding of developmental processes and disease progression. The platform’s user-friendly interface and integration with other bioinformatics tools make it accessible to both biologists and data scientists. Its modular design allows for the seamless incorporation of new trajectory inference methods, fostering continuous innovation in the field.
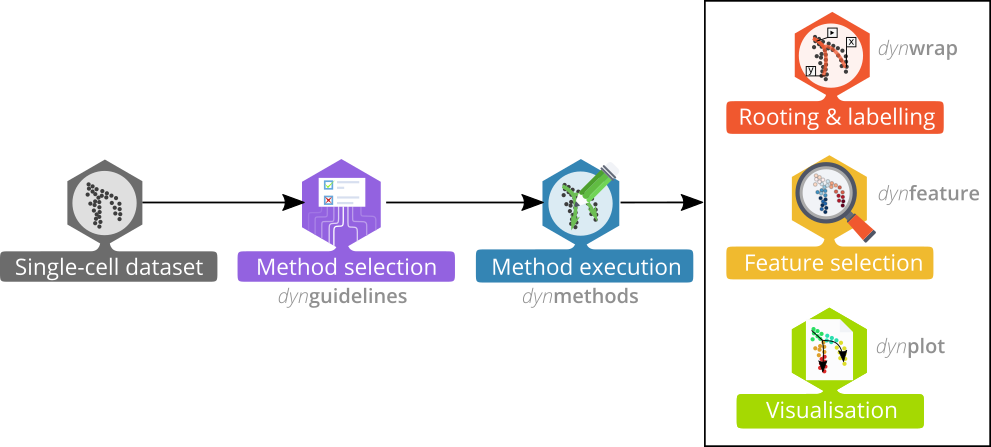
The dyno package offers end-users a complete TI pipeline. It features: - a uniform interface to 60 TI methods, - an interactive guideline tool to help the user select the most appropriate method, - streamlined interpretation and visualisation of trajectories, including colouring by gene expression or clusters, and - downstream analyses such as the identification of potential marker genes.
For information on how to use dyno, check out the installation instructions, tutorials and documentation at dynverse.org
Recent changes in dyno 0.1.1 (06-04-2019)
BUGFIX: Fixed example dataset
MINOR CHANGE: Bumping version requirement of dynmethods to 1.0.1.
Recent changes in dyno 0.1.0 (29-03-2019)
- Initial beta release of dyno
Dynverse dependencies